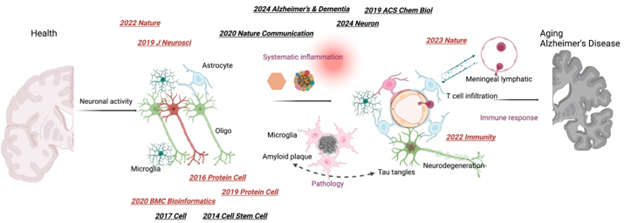
Carling G, Fan L, Foxe N, Norman K, Wong M, Zhu D, Corona C, Razzoli A, Yu F, Yarahmady A, Ye P, Chen H, Huang Y, Amin S, Sereda R, Lopez-lee C, Zacharioudakis E, Chen X, Xu J, Cheng F, Gavathiotis E, Cuervo A, Holtzman D, Mok S, Sinha S, Sidoli S, Ratan R, Luo W, Gong S, Gan L. Alzheimer’s disease-linked risk alleles elevate microglial cGAS-associated senescence and neurodegeneration in a tauopathy model. 2024. Neuron. 112. 1-20, PMID: 38328219
AAIC, Kloske C, Tansey M, Wilcock D (Chen X, leading the study of Microglia-mediated T-cell infiltration and reactivity) Advancements in Immunity and Dementia Research: Highlights from the 2023 AAIC Advancements: Immunity Conference. 2024. Alzheimer’s and Dementia. PMID: 39692624
AAIC, Kloske C, Bu G, Goate A, Holtzman D (Chen X, leading the study of APOE and immune response in Alzheimer’s Disease) Advancements in APOE and dementia research: Highlights from the 2023 AAIC Advancements: APOE conference. 2024. Alzheimer’s and Dementia. 20, 5815-6664, PMID: 39031528
Chen X, Firulyova M, Manis M, Herz J, Smirnov I, Aladyeva E, Wang C, Bao X, Finn M, Hu H, Shchukina I, Kim M, Yuede C, Kipnis J, Artyomov M, Ulrich J, Holtzman D. Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy. 2023. Nature. 615, 668-677, PMID: 36890231
Chen X, Holtzman D. Emerging roles of innate and adaptive immunity in Alzheimer’s Disease. 2022. Immunity. 55, 2236-2254, PMID: 36351425
II-Genetic and epigenetic regulations in learning in adult mammalian brain
Cellular diversification is critical for specialized functions of the brain including learning and memory. Single-cell RNA sequencing facilitates transcriptomic profiling of distinct major types of neurons, but the divergence of transcriptomic profiles within a specific neuronal population and their link to function remain poorly understood. Using single cell multi-omics, in vivo calcium imaging, optogenetics, CRISPR knockout, and behavior analyses, I discovered a subpopulation neuron that specifically undergoes transcriptomic plasticity in response to neuronal activity and learning. Those findings answer the fundamental question of how diversification of neurons influences learning and memory and opens an entirely new and exciting direction of research in neuroscience for studying functional diversity in subtypes of neurons in response to environmental changes.
Chen X, Du Y, Broussard J, Kislin M, Yuede C, Zhang S, Dietmann S, Gabel H, Zhao G, Wang S, Zhang X, Bonni A. Transcriptomic mapping uncovers Purkinje neuron plasticity driving learning. 2022. Nature. 605, 722-727, PMID: 35545673
Chen X, Zhang B, Wang T, Bonni A, Zhao G. Robust principal component analysis for accurate outlier detection in RNA-Seq data. 2020. BMC Bioinformatics. 21, 269. PMID: 32600248
Chen X, Chanda A, Ikeuchi Y, Zhang X, Goodman JV, Reddy NC, Majidi SP, Wu DY, Smith
SE, Godec A, Oldenborg A, Gabel HW, Zhao G, Bonni S, Bonni A. Transcriptional regulator SnoN promotes proliferation of cerebellar granule neuron precursors in the postnatal mouse brain. 2019. J Neurosci. 39, 44-62. PMID: 30425119
Smith SE, Chen X, Brier L, Bumstead J, Rensing N, Epstein A, Oldenborg A, Crowley J, Bice
A, Dikranian K, Ippolito J, Haigis M, Papouin T, Zhao G, Wong M, Culver JP, Bonni A.
Astrocyte deletion of α2-Na/K ATPase triggers episodic motor paralysis in mice via a metabolic pathway. 2020. Nature Communication. 11, 6164. PMID: 33268780
Krishnan N, Chen X, Donnelly-Roberts D, Mohler E.G, Holtzman D. M, Gopalakrishnan, S. M. Small molecule phenotypic screen identifies novel regulators of LDLR expression. 2020. ACS Chem Biol.15, 3262-3274. PMID: 33270420
III-Immune signaling in neuronal maturation
I studied the molecular and cellular mechanisms governing neuropsychiatric disorders by using functional subtype-specific neurons derived from human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs). I discovered that MeCP2-deficient human ESCs/iPSCs-derived neurons showed aberrant immature action potentials and synaptic transmission. To capture the molecular mechanisms governing neuron electrophysiological maturation at single cell level, I therefore developed electrophysiological recording coupled with single cell transcriptome analysis (Patch-Seq) and revealed a tight link between neuronal maturation and genes involved in ubiquitination, immune function, and oxidative phosphorylation.
Chen X*, Han X, Blanchi B, Guan W, Ge W, Yu YC*, Sun YE*. Graded and pan-neural disease phenotypes of Rett Syndrome linked with dosage of functional MeCP2. 2020. Protein Cell. 12, 639-652. PMID: 32851591 (co-corresponding author)
Chen X, Zhang K, Zhou L, Gao X, Wang J, Yao Y, He F, Luo Y, Yu Y, Li S, Cheng L, Sun YE.
Coupled electrophysiological recording and single cell transcriptome analyses revealed molecular mechanisms underlying neuronal maturation. 2016. Protein Cell. 7, 175-186. PMID: 26883038 (Cover research)
Chen Y, Yu J, Niu Y, Qin D, Liu H, Li G, Hu Y, Wang J, Lu Y, Kang Y, Jiang Y, Wu K, Li S, Wei J, He J, Wang J, Liu X, Luo Y, Si C, Bai R, Zhang K, Liu J, Huang S, Chen Z, Wang S, Chen X, Bao X, Zhang Q, Li F, Geng R, Liang A, Shen D, Jiang T, Hu X, Ma Y, Ji W, Sun YE. Modeling Rett Syndrome Using TALEN-Edited MECP2 Mutant Cynomolgus Monkeys.
2017. Cell. 169, 945-955. PMID: 28525759
Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S, Jing B, Si C, Lin Q, Chen X, Lin H, Pu X, Wang Y, Qin B, Wang F, Wang H, Si W, Zhou J, Tan T, Li T, Ji S, Xue Z, Luo Y, Cheng L, Zhou Q, Li S, Sun YE, Ji W. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. 2014. Cell Stem Cell. 14, 323-328. PMID: 24529597
Research Accomplishment
-Ten years scientific journey in filling the gap of neuron-immune interactions